Tags: [Bio] - dynamic environments, eco/evolution, information processing, [Learning] - learning without neurons in molecules + mechanics
2024 - 2022
|
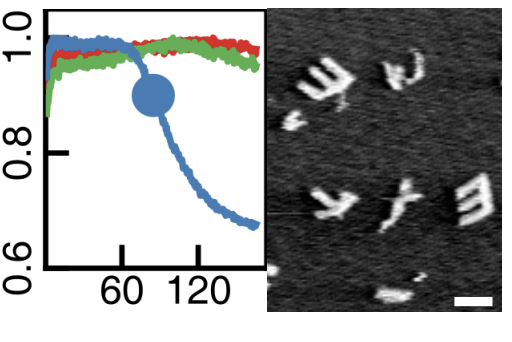
Pattern recognition in the nucleation kinetics of non-equilibrium self-assembly [Bio] [Learning]
C.G. Evans, J.O. Brien, E. Winfree, A. Murugan Nature (2024) Can a soup of molecules act like a neural network? We experimentally demonstrate a molecular system here that acts like an associative neural network - similar architecture, pattern recognition behavior, ability to expand in a Hebbian manner, phase diagrams etc. But we didn't design these molecules to act like `neurons' in any sense - they are just molecules following the inevitable physics of nucleation, self-assembly and depletion. You can easily imagine such hidden computational power in molecular collectives being exploited by evolution. Lesson: Neural computation doesn't need networks of linear threshold devices (`neurons'); it can arise through the collective dynamics of other many-body systems. |
|
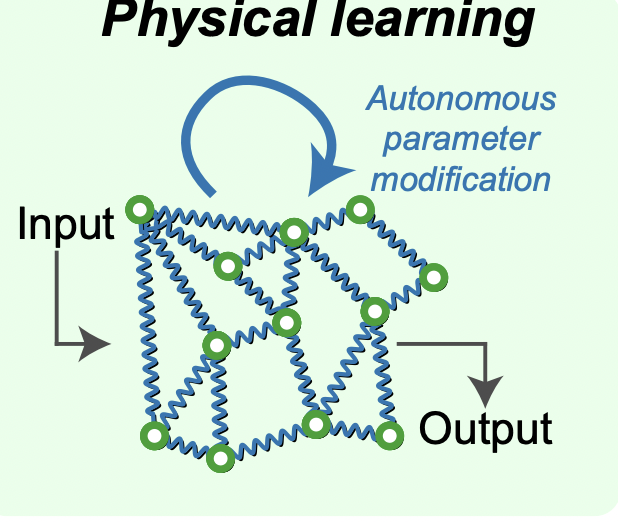
Learning without neurons in physical systems [Learning]
N. Stern, A. Murugan Annual Reviews of Condensed Matter Physics (2023) We review the incipient field of `physical learning' - when can a physical system physically learn desired behaviors by experiencing examples of such behavior? We categorize examples in the molecular and mechanical literature as unsupervised or supervised training. We highlight the main intellectual challenges - e.g., how can learning work despite learning rules having to be local in space and time? What does all of this have to do with adjacent mature fields like biological learning and neuromorphic computing? |
|
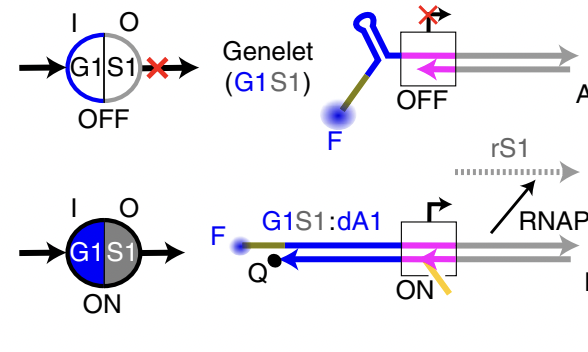
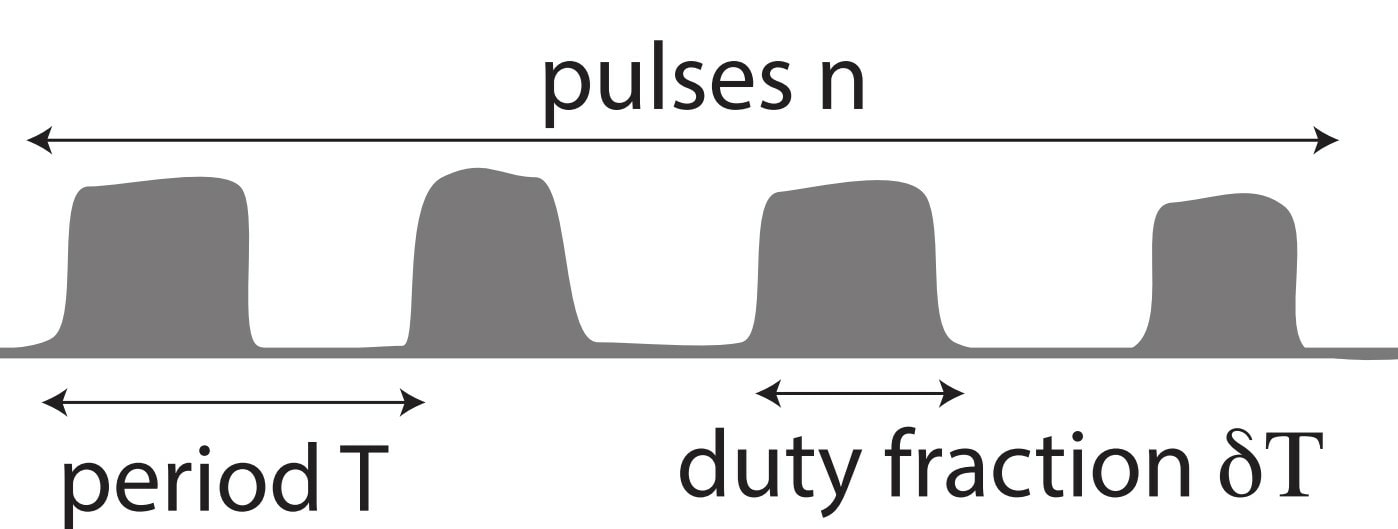
Standardized excitable elements for scalable engineering of far-from-equilibrium chemical networks, [Learning]
S W. Schaffter, K-L Chen, J O’Brien, M Noble, A Murugan, R Schulman* Nature Chemistry (2022) Sam Schaffter and others in Schulman's lab created an amazing scaleable platform for robust synthetic molecular circuits based on `genelets' (little modules of DNA + RNA + a RNA polymerase). Today, these elements can already be linked together to create multi-stable systems, generate pulses and the like. Tomorrow, maybe these chemical circuits can be combined with mechanical systems in Schulman's lab to reveal a whole new class of materials that can learn a la neural networks. |
|
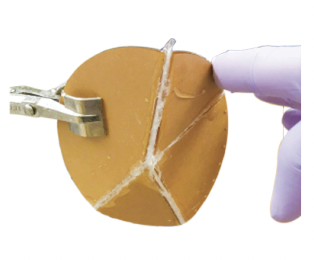
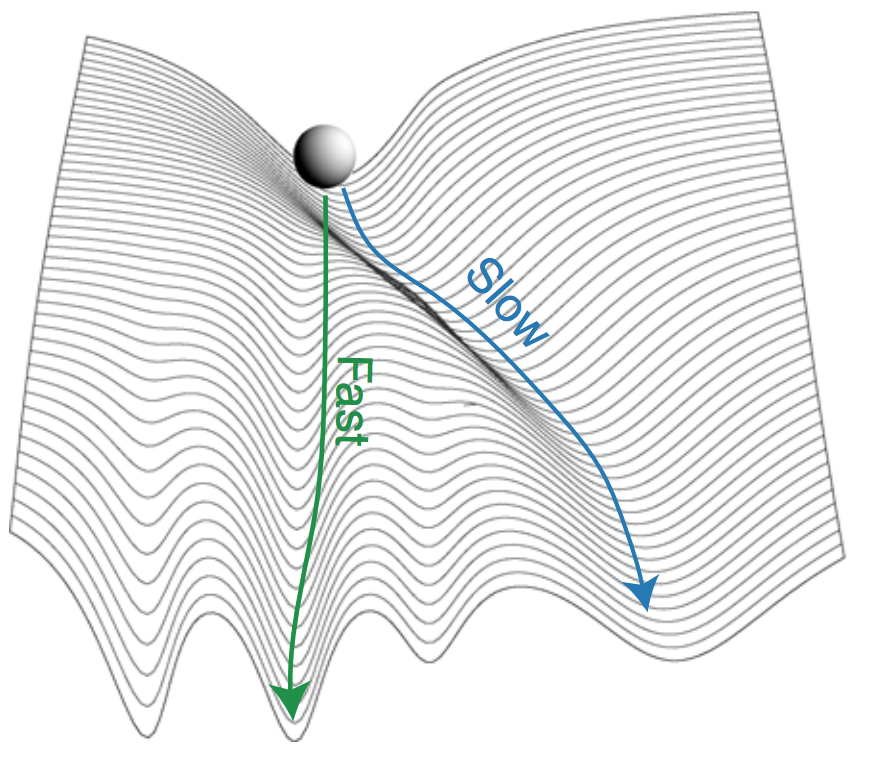
Learning to self-fold at a bifurcation [Learning]
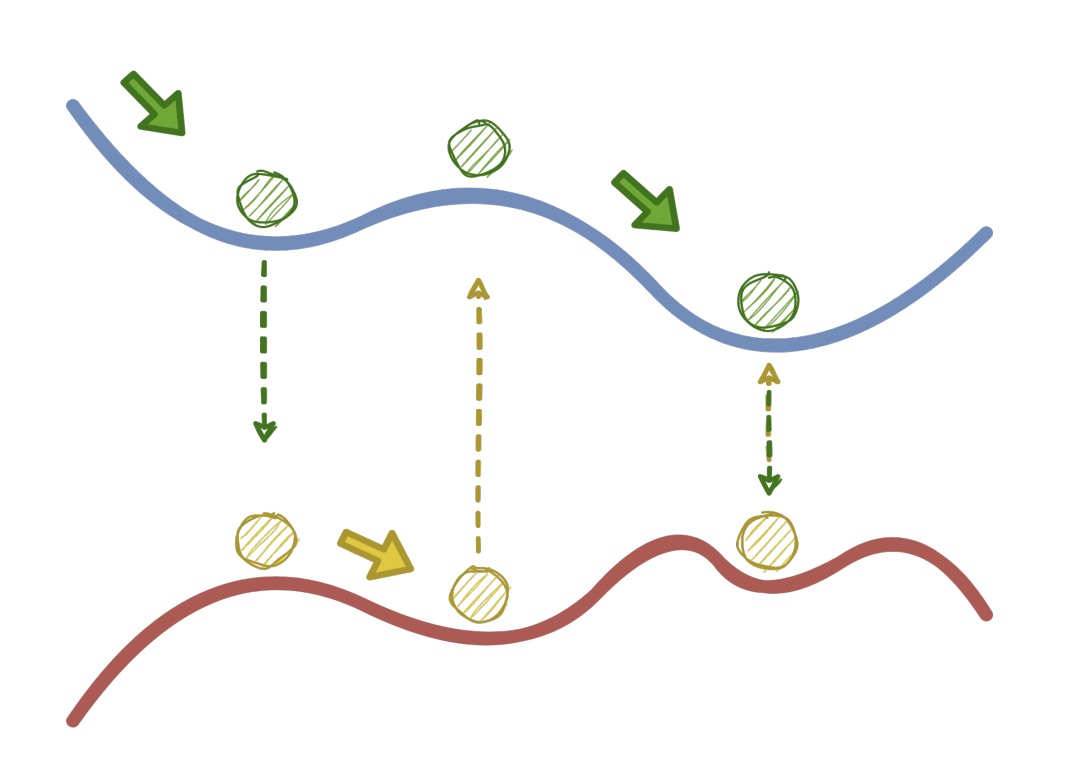
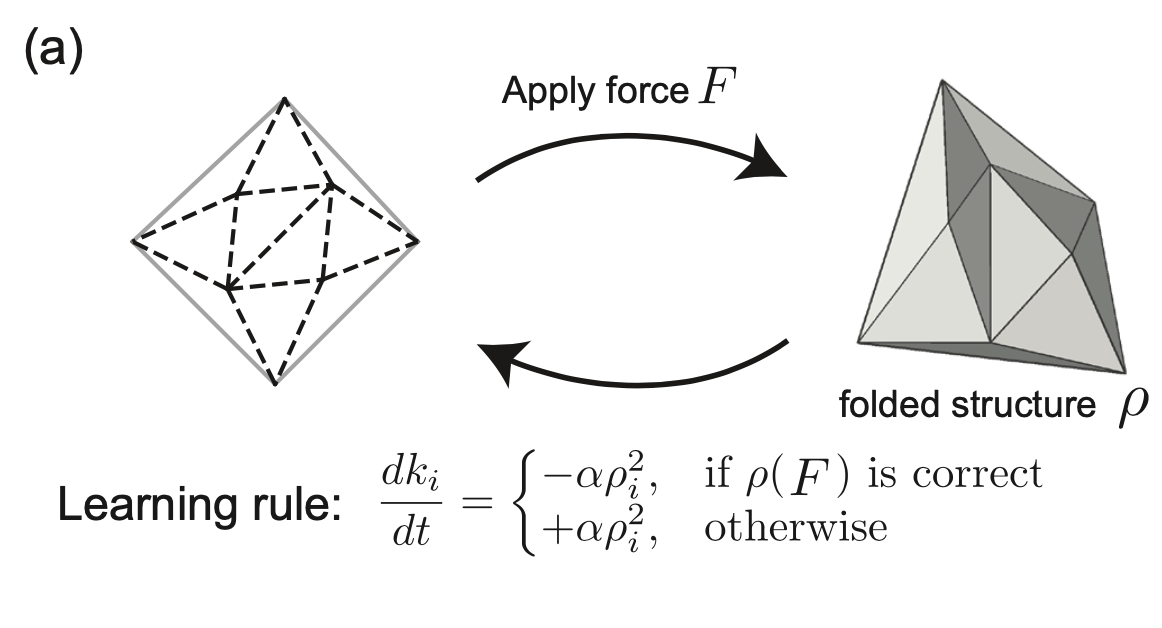
Arinze C, Stern M, Nagel SR, Murugan A. Physical Review E (2023) Bifurcations are special states in mechanical systems where any linear approximation completely misses the picture, even for small deformations. How do we design the behavior near these highly non-linear points? We demonstrate experimentally that mechanical systems can `physically learn' desired behaviors at these points. We fill a creased sheet with soft epoxy and physically subject it to the desired behavior as the epoxy is setting; we find that the epoxy re-distributes itself in just the right way so as to learn the desired behavior at the bifurcation. No computers or design algorithms involved! |
|
Non-Convex Optimization by Hamiltonian Alternation. [Bio] [Learning]
A Apte, K Marwaha, A. Murugan, arXiv (2022) Multitasking is distracting and often makes you terrible at each of your tasks. But sometimes, if you are stuck on a given task, switching to a different task for a while can help you get unstuck. But how do you know when this will work and what kind of task should you switch to? If the `task' is finding the ground state of a Hamiltonian, we have one simple (analytic) trick to get unstuck - we found an analytically-specified simple alternative Hamiltonian that you should switch to every so often during minimization. Guaranteed* to help you get unstuck (*conditions apply). |
2021
|
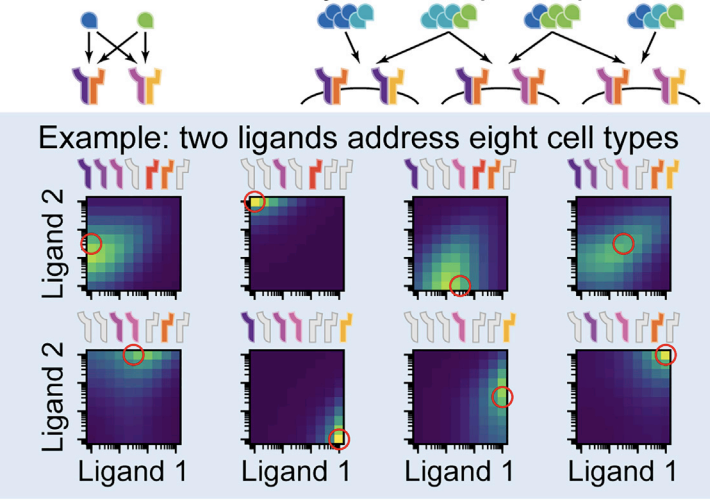
Ligand-receptor promiscuity enables cellular addressing. [Bio]
C Su, A Murugan, J Linton, A Yeluri, J Bois, H Klumpe, Y Antebi, M Elowitz Cell Systems 2021 Criss-crossed phone lines sound like a bad idea if you want to communicate from one specific place to another (at least back when phones had lines). Can it ever make sense to connect phone lines from different houses into one messy ball of wires and then fan them out to different destinations? It turns out that molecules can handle such a messy ball just fine - the collective dynamics of these molecules will eventually sort out messages to the right destinations. In fact, there are advantages to being this messy. |
|
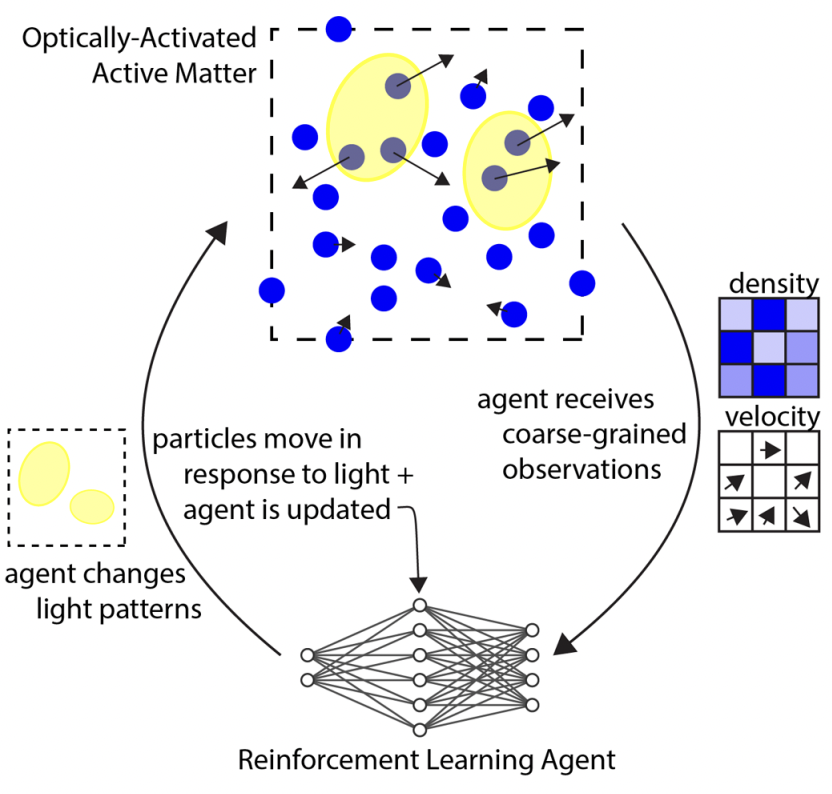
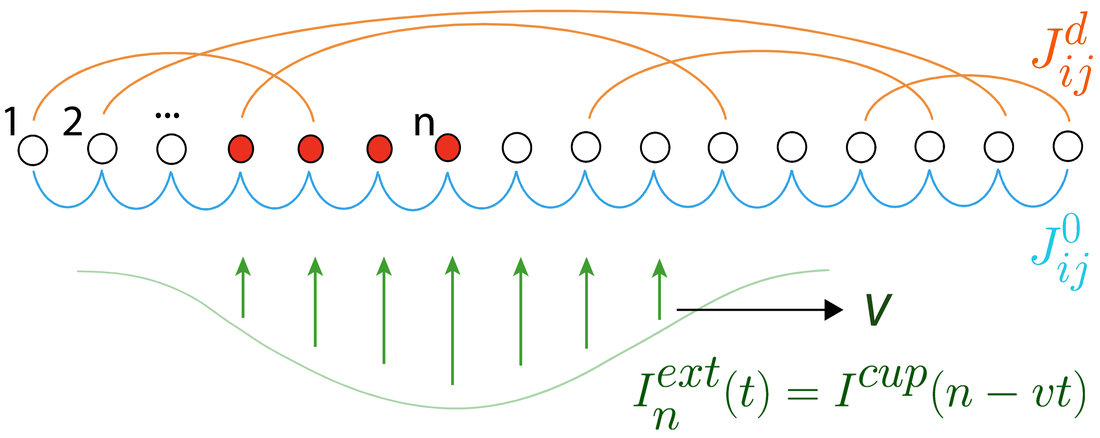
Learning to control active matter [Learning]
M. Falk, V. Alizadehyazdi, H. Jaeger, A. Murugan Physical Review Research (2021) Active matter is seen as a metaphor for biology because active matter is out of equilibrium. But lots of physical phenomena are out of equilibrium and most are boring. What makes biological phenomena uniquely interesting is the ability to regulate the flow of energy in functional ways. Here, we ask if regulating activity in a space-time dependent way can create interesting organization in a minimal active matter systems. |
|
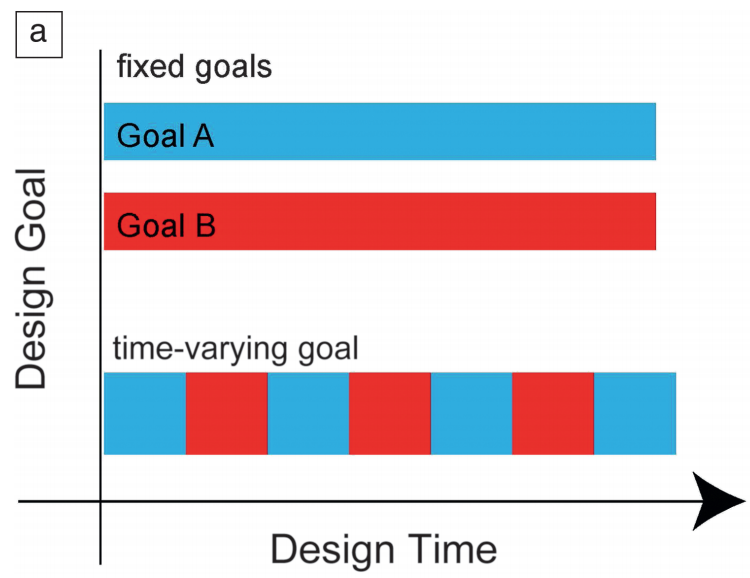
Roadmap on biology in time varying environments [Bio]
A. Murugan et al Physical Biology 2021 A review. Everyone talks about biology in time varying environments but only in some cases do time varying environments do something truly novel - there is no way to understand the resulting behavior in terms of any averaged or effective static environment. This review, by leaders in many distinct parts of biology, explores such conceptual questions in different areas + what the promising theoretical and experimental approaches are. |
|
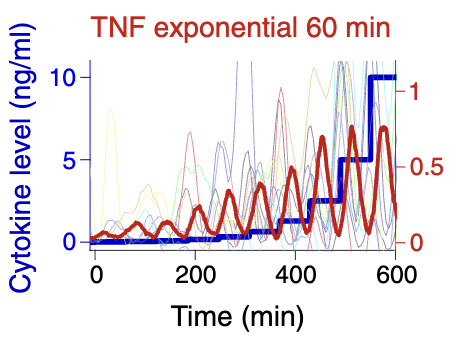
NF-κB responds to absolute differences in cytokine concentrations [Bio]
M Son, A G Wang, H Tu, M O Metzig, P Patel, K Husain, J Lin, A Murugan, A Hoffmann, S Tay Science Signaling 2021 Jan 19;14(666):eaaz4382 Molecular information isn't just in the chemical identity or amounts of molecules; sometimes, information is encoded in the pattern of how an amount changes over time. But while our brain can easily decode information in a time-varying signal (e.g., speech), how do molecular circuits perceive and decode such temporally coded information? Here is a case study where the circuit seems to respond to absolute differences and not to the baseline concentration. That requires both memory (to compute the difference) and the ability to forget (to ignore the baseline). |
2020
|
Physical constraints on epistasis [Bio]
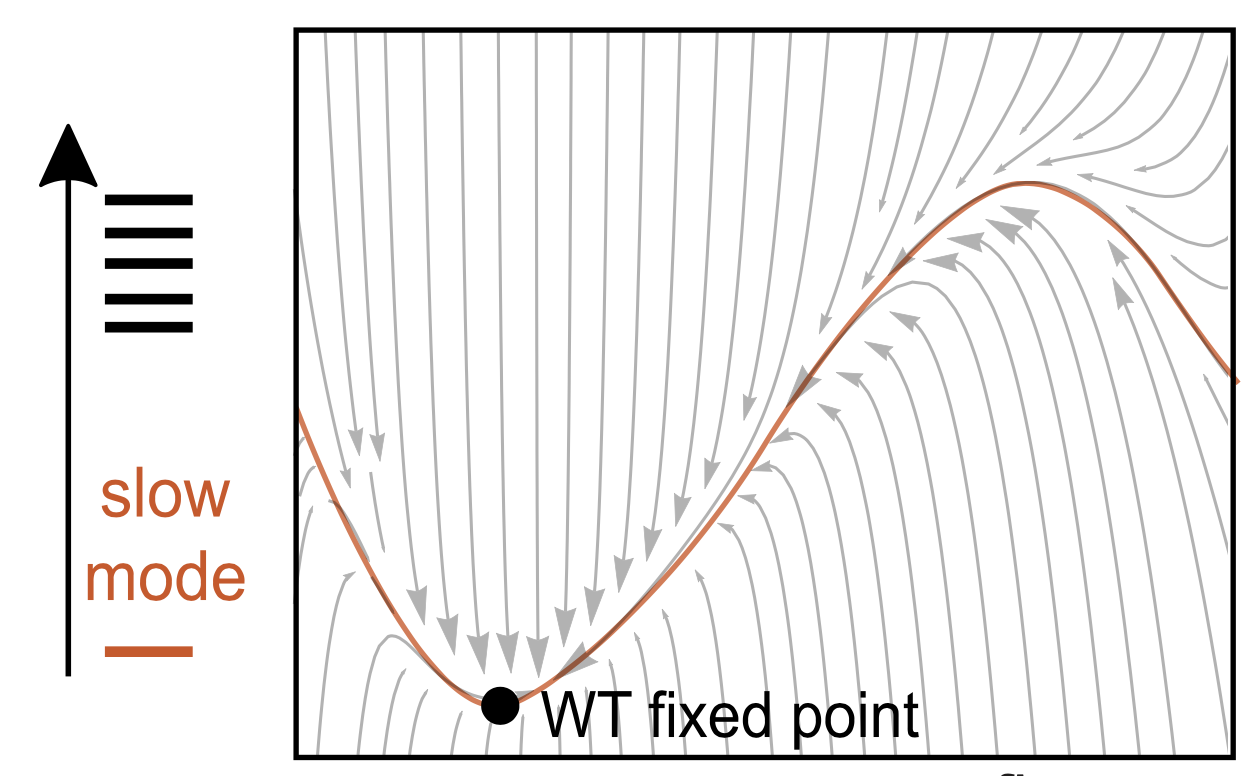
Molecular Biology and Evolution, 2020 K Husain, A Murugan The combined effect of two changes cannot usually be predicted from the effect of individual changes in a complex system. But people have noticed that mutational experiments in protein and across the genome are more predictable than expected. We suggest a mechanistic reason for such simplicity by exploiting a surprising connection - obvious, once you see it - between the physical dynamics of a system (perturbing the state) and its evolutionary dynamics (perturbing parameters). |
|
Supervised learning through physical changes in a mechanical system [Learning]
M. Stern, C. Arinze, L. Perez, S. Palmer, A Murugan PNAS, 2020 Can a mechanical system be physically trained to show a behavior the way neural networks are trained to recognize images of cats and dogs? We show that a mechanical system can be trained to show the right physical response (analogous to shouting 'Dog!', 'Cat!') to a given force pattern (analogous to images of dogs/cats) by either softening or stiffening parts of the material in proportional to local strain, depending on whether the response was right or wrong. Most critically, our training rules are `local' in space, so real physical processes can easily implement such dynamics. No computers involved! |
|
Tuning environmental timescales to evolve and maintain generalists [Bio]
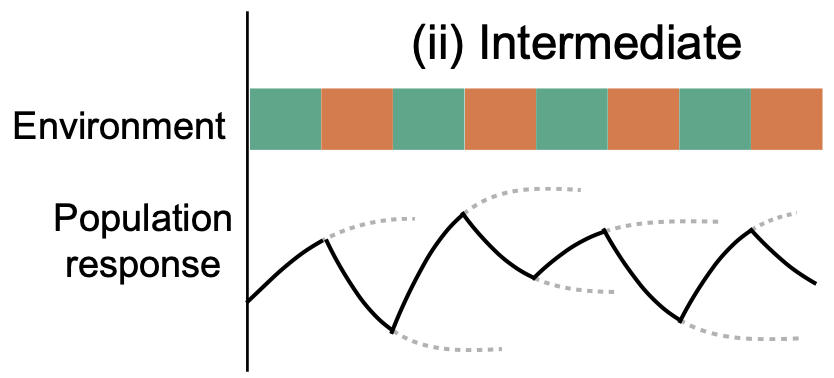
V Sachedva*, K Husain*, J Sheng, S Wang+, A Murugan+ PNAS, 2020 When can your immune system learn the right lessons from past exposure to viruses? Learning too much is bad - viruses mutate and the virus of tomorrow will be different from the variant you see today. We found that highly dynamic exposure to different viral mutants over time prevent you from learning the wrong details. E.g., alternating exposure to different viral mutants in a faster and faster sequence - a "chirp" - gives you pretty good odds of learning the right general lessons. |
|
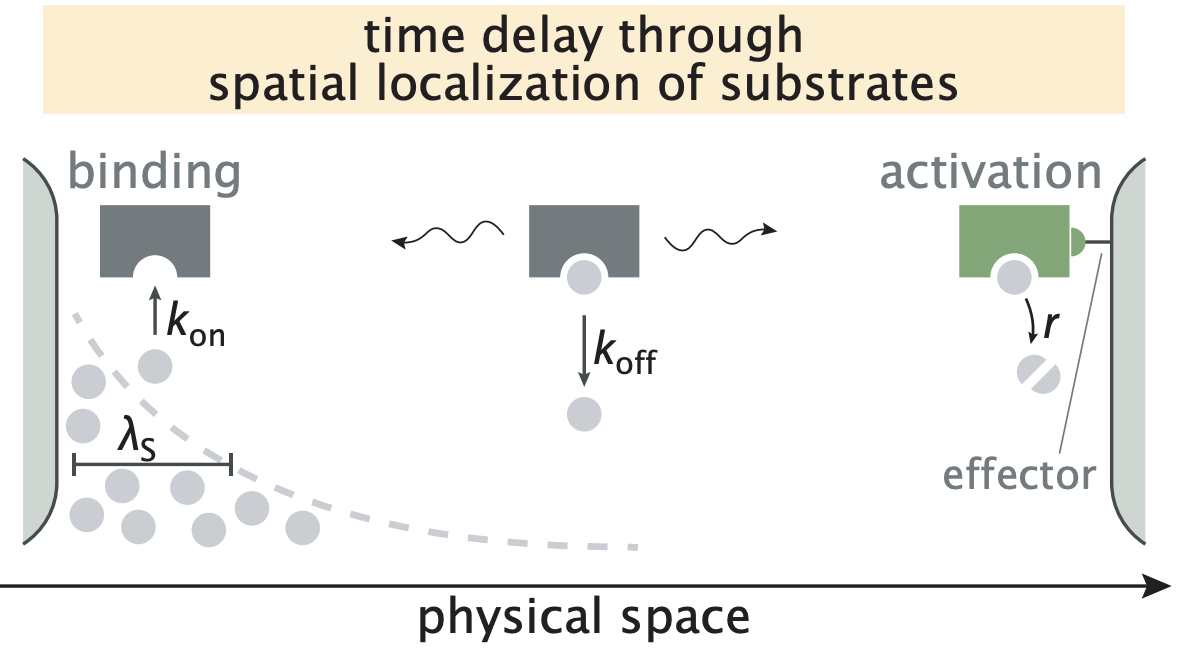
Proofreading through spatial gradients [Bio]
V. Galstyan, K. Husain, F. Xiao, A. Murugan+, R. Phillips+ eLife, 2020 John von Neumann, Ralf Landauer and Charlie Bennett showed that information processing costs energy. John Hopfield showed a remarkable consequence of this "Landauer principle" in biology - some biological enzymes have evolved complex features to consume chemical energy in order to make fewer mistakes than equilibrium thermodynamics requires. We argue that enzymes without any chemical energy available can still perform such non-equilibrium information processing by exploiting the fact that the cell is not well-mixed. Moral: spatial structure is intrinsically out of equilibrium and can be an alternative to chemical energy. |
|
Continual learning of multiple memories in mechanical networks. [Learning]
M. Stern, M. Pinson, A. Murugan Physical Review X, 2020 Many neural networks cannot learn new things in a sequential manner because the synaptic connections formed to learn the new things might interfere with retrieving older memories. Here, we ask when an elastic network, grown over time, can recall the different geometries it experienced. We find that mechanical properties constrain information processing and memory capacity - e.g., non-linear springs are required for reasons deeply connected to non-linear priors used in sparse regression. |
|
Non-equilibrium statistical mechanics of continuous attractors [Bio] [Learning]
W. Zhong, Z. Lu, D.J.Schwab+, A. Murugan+ Neural Computation (2020) Recent experiments suggest that memories of multiple spatial environments involve the same neurons in the brain. Why do we not get confused? Classic "equilibrium" theories do not apply to such spatial memories (`continuous attractors'). We compute the speed-dependent "non-equilibrium" memory capacity of a neural network and show that the faster you run, the less you remember. |
2019
|
Kalman-like self-tuned sensitivity in biophysical sensing. [Bio]
K Husain, W Pittayakanchit, G Pattanayak, M J Rust, A. Murugan Cell Systems 2019, 9(5) P459-465.E6 People often view biological systems through the lens of a physical system with fixed parameters and speak of trade-offs, e.g., you cannot be both fast and accurate. But biological systems can tune most parameters over time based on how things are working out for them. Using experiments on circadian clocks in cyanobacteria (from Prof. Rust's lab at Chicago) and on stress response pathways in yeast (from Prof. Swain's lab in Edinburgh), we showed that biological systems are like dynamical systems whose geometry is changed over time in response to the system's performance. As a result, living organisms can self-tune their sensitivity to new external information, a foundational idea in engineering proposed by Kalman (1960). |
|
Bioinspired nonequilibrium search for novel materials. [Learning]
A. Murugan, H. Jaeger MRS Bulletin 44(2):96-105 (2019) pdf here Non-equilibrium physics + bio-materials are all the rage these days. In this perspective, we emphasize a distinct but profound role for non-equilibrium physics --- the non-equilibrium dynamics of evolutionary processes that produced at such materials in the first place. Understanding non-equilibrium evolutionary dynamics will tell us how to design materials to be more adaptable like biomaterials. |
|
Temporal pattern recognition through analog molecular computation. [Bio] [Learning]
with: J O'Brien ACS Synthetic Biology (March 2019) Popular summary by MIT Tech Review Chemists have long made sensors to detect a specific molecule and raise an alarm. But what if we need to raise an alarm only for a specific pattern of exposure to a molecule over time? Can the internal dynamics of a sensor extract specific features of a time-varying signal? |
2018
|
Shaping the topology of folding pathways in mechanical systems. [Learning]
M. Stern, V. Jayaram, A. Murugan Nature Communications 9:4303 (2018) When pressured, elastic materials sometimes face a fork in the road and they take it. But often, we'd rather not give them the choice. We found that stiffening some parts of the material can cause `bifurcations' that relocate or even remove forks, so the network of roads only connects places you like. Even better, some roads are accessible only at 60 mph while others accessible only at 20 mph. |
|
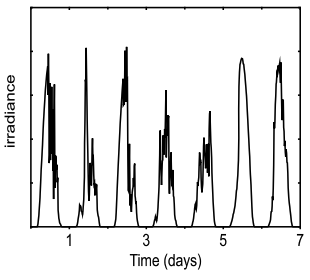
Information content of downwelling skylight for non-imaging visual systems (bioRxiv, Sep 2018) [Bio]
R. Thiermann, A. Sweeney, A. Murugan Many organisms have light-sensitive proteins (opsins) in unexpected places like in the brain and in reproductive organs. What are these opsins "looking" at? We aren't sure but they seem to link reproductive behaviors to the lunar cycle. Using data on natural light from Prof. Sweeney and weather models, we find these opsins can reliably tell the day of the lunar cycle from the color of twilight. |
|
Biophysical clocks face a trade-off between internal and external noise resistance. [Bio]
W Pittayakanchit*, Z Lu*, J Chew, M J Rust, A. Murugan eLife (2018);7:e37624 Many organisms have 24-hour self-sustained internal clocks but other organisms have simpler `hourglasses' turned over by sunrise and sunset. We study the geometry of the underlying dynamical systems and find that self-sustained clocks are better at dealing with external weather fluctuations. But hourglasses are better at dealing with internal molecular fluctuations. So a simple hourglass can be better than a fancy clock when the clock hardware itself is error prone. High Protein Copy Number Is Required to Suppress Stochasticity in the Cyanobacterial Circadian Clock [Bio] J. Chew, E. Leypunskiy, J. Lin, A. Murugan, M. Rust Nature Communications 9:3004 (2018) Related experiments in Prof. Michael Rust's lab show that P. marinus, a cyanobacterium with an hourglass clock, has very few clock proteins compared to a sister species S. elongatus that has a self-sustained clock. These experiments also suggest that P. marinus is better off with a simple hourglass clock than with a self-sustained clock, given how unreliable the clock reactions in P. marinus are. |
2017
|
Topologically protected modes in non-equilibrium stochastic systems. [Bio]
A. Murugan, S. Vaikuntanathan Nature Communications 8, 13881 (2017) Sometimes, every little change to parameters changes how a system behaves. But sometimes, the behavior barely changes. People have identified a large class of linear systems where the latter can be interpreted in terms of conserved integers (`topological protection'). We extended this class to include non-Hermitian matrices that describe Markov chains. |
|
The Complexity of Folding Self-Folding Origami. [Learning]
M. Stern, M. Pinson, A. Murugan Physical Review X 7, 041070 (2017) The dream of self-folding origami is to program a flat sheet with carefully placed creases so the sheet folds itself into a swan when pushed anywhere. We show that, much like with protein folding, the very act of programming a sheet to fold in one way (e.g., a swan) also lets it fold into an exponential number of other undesired shapes. Folding such a sheet is as difficult as solving a Suduko puzzle. |
|
Associative pattern recognition through macro-molecular self-assembly. [Bio] [Learning]
W. Zhong, D.J. Schwab, A. Murugan Journal of Statistical Physics, Kadanoff memorial issue (2017) Can self-assembling molecules act like a convolutional neural network? We propose a setup that can distinguish subtle patterns in the concentrations of several molecules that correspond to corrupted images of Leo Kadanoff, a cat and Albert Einstein. The self-assembling setup naturally shows large translational invariance like a convolutional neural network. |
|
Self-folding origami at any energy scale. [Learning]
M. Pinson*, M. Stern*, A. Carruthers, T. Witten, E. Chen, A. Murugan Nature Communications 8:15477 (May 2017) "Mechanisms" are motions in mechanical systems that cost exactly zero energy. Clever people like James Watt have designed elaborate mechanisms that have changed the world. But physics does not sharply distinguish "exactly zero energy" vs nearly zero energy. We go beyond "exactly zero energy" and describe the "typical" folding motions at each energy. |
2016
|
The Information Capacity of Specific Interactions. [Bio] [Learning]
M. Huntley*, A. Murugan* , M. Brenner, Proceedings of the National Academy of Sciences (2016) Shannon used information theory to quantify how many distinct messages you can communicate over a noisy cable. We use information theory to quantify how many distinct physical components (molecules, colloids etc) can be distinguished by the physics of the system (DNA hybridization, depletion interactions etc). We find that each physical system has a Shannon capacity - throw in any more components than the capacity and the components can't tell each other apart anymore. |
|
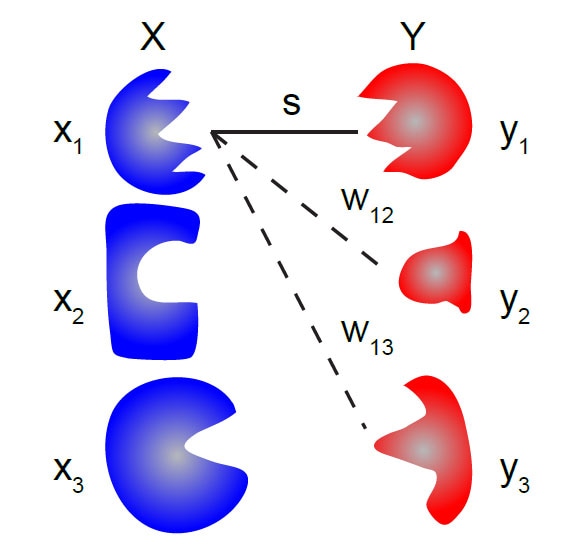
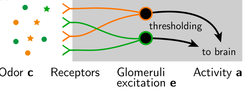
Receptor libraries optimized for natural statistics. [Bio]
D. Zwicker, A. Murugan*, M. Brenner*, (* corresp. authors) Proceedings of the National Academy of Sciences (2016) When you play a game of 20 questions, the optimal questions to ask each divide the set in half and are `orthogonal' to each other. For example 'Are you thinking of a man or a woman?', 'Is the person dead or alive?' and so on. What is optimal set of 20 questions to ask of odors in your natural environment? The answer provides design principles for the set of olfactory receptors in your nose. |
|
Biological implications of dynamical phases in non-equilibrium reaction networks, [Bio]
A. Murugan, S. Vaikuntanathan invited contribution to the Journal of Statistical Physics (special issue), 2016, 162 (5) JSP arXiv Many biochemical mechanisms use non-equilibrium driving to dramatically change the state in which a bio-molecular system spends most of its time when biologically desired but not otherwise. In this review, we relate such abilities, which underly biochemical error correction and adaption, to the idea of non-equilibrium dynamical phases. We find that dynamical phase coexistence creates special `common-sense' points in the energy-accuracy tradeoff where you achieve 80% of your goals with 20% effort. |
2015
|
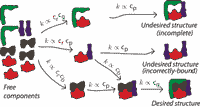
Undesired usage and the robust self-assembly of heterogeneous structures, [Bio] [Learning]
in collaboration with: J. Zou, and M. Brenner Nature Communications 6, 6203 (Jan 2015) Nat Comm. Correct self-assembly of structures made of many distinct species must compete against a combinatorially enormous number of undesired structures. However, these entropic challenges can be overcome to a remarkable extent by tuning control parameters (like concentrations) to reflect the 'undesired usage' of species. That is, you must set your control knobs based on *undesired* structures, instead of the desired structure as usually assumed. Hence, somewhat counterintuitively, highly non-stoichiometric concentrations can greatly enhance the yield of desired structures. |
2014
|
Multifarious Assembly Mixtures: Systems Allowing Retrieval of Diverse Stored Structures, [Bio] [Learning]
in collaboration with: Z. Zeravcic, S. Leibler and M. Brenner Proceedings of the National Academy of Sciences 112(1) 54-59 (Dec 2014) arXiv PNAS Inspired by associative memory in Hopfield's neural networks, we generalized the self-assembly framework to a soup of particles (proteins/DNA tiles) that can simultaneously 'store' the ability to assemble multiple different structures. Such a soup of particles can then assemble ('retrieve') any one of the stored structures ('memories') when presented with a signal vaguely reminiscent of one of those memories ('association'). However, store one too many memories and promiscuous interactions between particles prevent faithful retrieval of any memory. Secretly, such self-assembly is conceptually similar to Hippocampus place cell networks and equivalent spin glass models. |
|
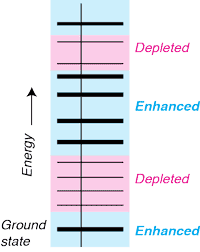
Discriminatory proofreading regimes in non-equilibrium systems, [Bio]
in collaboration with: D.A. Huse, and S. Leibler Physical Review X 4 (2), 021016 PRX Usually, non-equilibrium error correction is understood to increase the occupancy of the ground state and reduce the occupancy of all higher energy states. However, we found that a proofreading mechanism can act differently in different energy bands, reducing occupancy of unstable states in a given energy band while increasing the occupancy of less stable higher energy states ('anti-proofreading'). And you can switch between different designer occupancy of states by simply changing the external driving forces. |
2013
|
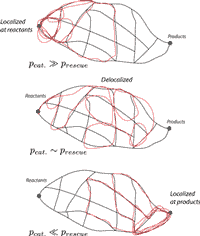
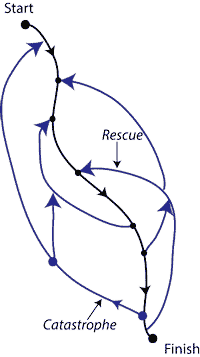
Speed, dissipation, and error in kinetic proofreading, [Bio]
in collaboration with: D.A. Huse, and S. Leibler Proceedings of the National Academy of Sciences 109(30):12034-9 (2012) PDF A new twist on the classic model of kinetic proofreading. Proofreading uses 'catastrophes' to slow down biochemical reactions while improving their fidelity. We introduced `rescues' that mitigate catastrophes and speed up reactions at the cost of increased errors. Surprisingly, we found a non-equilibrium phase transition as you tune the rescue rate. At this transition, you achieve, loosely speaking, 80% of the max possible error-correction at only 20% of the max speed cost. Why would you go any further (as the traditional limit does) unless you really care about errors and really don't care about speed at all? We took the terms catastrophes and rescues from non-equilibrium microtubule growth. The connection to 'dynamic inability' of microtubules suggests a broader context for proofreading as a stochastic search strategy, balancing exploration and exploitation. |
Older: High energy physics
|
AdS4/CFT3 - squashed, stretched and warped,
in collaboration with: I.R.Klebanov and T.Klose Journal of High Energy Physics 0903 140 (2009) arxiv:0809.3773 [hep-th] PDF In its earliest form, the AdS/CFT correspondence related a gravitational theory on an Anti-de Sitter space \times a perfect sphere to the most supersymmetric conformal quantum field theory -- which has very little physics left because of all the symmetries. What happens if you take the perfect 7-dimensional round sphere of the gravitational theory and squash it, stretch it and then also warp it? |
|
Goldstone Bosons and Global Strings in a Warped Resolved Conifold,
in collaboration with: I. R. Klebanov, D. Rodriguez-Gomez and J. Ward Journal of High Energy Physics 0805, 090 (2008), arXiv:0712.2224 [hep-th] PDF The AdS/CFT correspondence relies heavily on matching symmetries of a gravitational theory with the symmetries of a quantum field theory. What happens when some of these symmetries are spontaneously broken? What do Goldstone modes of the field theory correspond to on the gravitational side? |
|
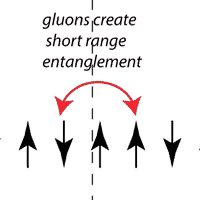
Entanglement as a Probe of Confinement,
in collaboration with: I. R. Klebanov and D. Kutasov Nuclear Physics B 796, 274 (2008), arXiv:0709.2140 [hep-th] PDF Entanglement entropy was traditionally not used much in high-energy physics (outside of black hole physics). We proposed a concrete use of entanglement entropy in particle physics - as a test of whether quarks are confined by gluons. Quark confinement is a basic feature of the real world but it can surprisingly difficult and subtle to check if a proposed theory of quarks really shows confinement. Entanglement entropy provides a tractable way. |
|
Gauge/Gravity Duality and Warped Resolved Conifold,
in collaboration with: I. R. Klebanov Journal of High Energy Physics 0703, 042 (2007), arXiv:hep-th/0701064 PDF Conical singularities in Einstein's theory of gravitation can be `resolved' away (in the sense of algebraic resolutions); the singularity is then replaced with a sphere of smaller dimension. We worked out what such resolution means for dual quantum field theories. |
|
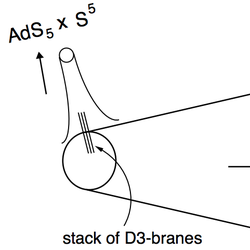
On D3-brane potentials in compactifications with fluxes and wrapped D-branes
in collaboration with: D. Baumann, A. Dymarsky, I. R. Klebanov, J. M. Maldacena and L. P. McAllister Journal of High Energy Physics 0611, 031 (2006), arXiv:hep-th/0607050 PDF String theorists wanted to build models of cosmic inflation based on string theory, which usually amounts to a balling rolling down a potential and emitting gravitational waves. However, such calculations for potentials obtained through string theory were too difficult. We made these calculations easy for a wide class of models by outlining a geometric method. |
|
Fatgraph expansion for noncritical superstrings, in collaboration with: Anton Kapustin PDF arXiv:hep-th/0404238 (2004) We studied a fatgraph expansion for noncritical strings. What more can I say? |